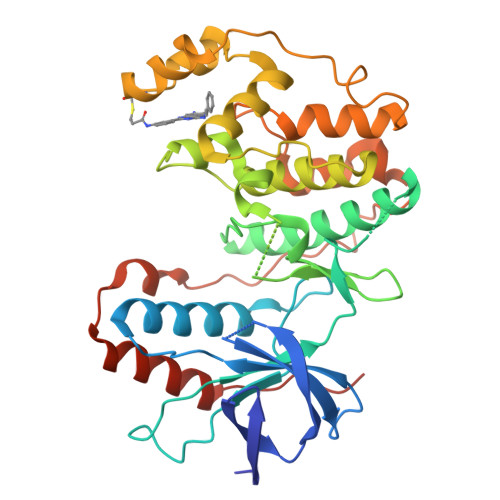
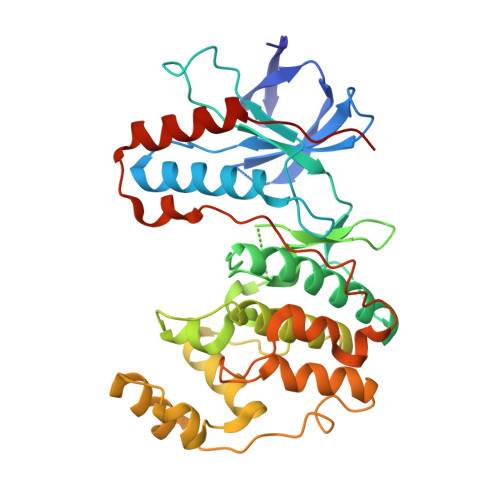
Covalent Lipid Pocket Ligands Targeting p38 alpha MAPK Mutants.
Buhrmann, M., Hardick, J., Weisner, J., Quambusch, L., Rauh, D.(2017) Angew Chem Int Ed Engl 56: 13232-13236
- PubMed: 28834017
- DOI: https://doi.org/10.1002/anie.201706345
- Primary Citation of Related Structures:
5O8U, 5O8V - PubMed Abstract:
A chemical genetic approach is presented to covalently target a unique lipid binding pocket in the protein kinase p38α, whose function is not yet known. Based on a series of cocrystal structures, a library of 2-arylquinazolines that were decorated with electrophiles were designed and synthesized to covalently target tailored p38α mutants containing artificially introduced cysteine residues. Matching protein-ligand pairs were identified by MS analysis and further validated by MS/MS studies and protein crystallography. The covalent ligands that emerged from this approach showed excellent selectivity towards a single p38α mutant and will be applicable as suitable probes in future studies of biological systems to dissect the function of the lipid pocket by means of pharmacological perturbations.
Organizational Affiliation:
Faculty of Chemistry and Chemical Biology, TU Dortmund University, Otto-Hahn-Strasse 4a, 44227, Dortmund, Germany.